What I am primarily interested in, is the data format
.parquet. It is advertised as an open source,
column-oriented data file format designed for efficient data storage and
retrieval. Well, the column oriented seems to be aligned
with how R works.
See more info on the {arrow} package.
Want to learn a bit more for Apache Arrow and dplyr for exploratory data analysis, see this amazing presentation by Tom Mock
Last, the code for the entire analysis canbe found on github
Let’s code now.
First, the needed libraries.
The dataset
I am going to use the palmerpenguins dataset. It’s a dataset with 3 character columns and 5 numeric ones.
Will do the following
- Increase the row size to 100,000
- Turn factor columns to character
Write data to disk
We write a .csv version, and a .parquet
version
temp_file_csv <- tempfile(pattern = ".csv")
temp_file_parquet <- tempfile(pattern = ".parquet")
write.csv(dta, temp_file_csv, row.names = FALSE)
arrow::write_parquet(dta, temp_file_parquet)
The comparison
I use the {bench} package.
Note the check = FALSE argument in
bench::mark(). This argument checks whether the results of the different methods are consistent.
There is a reason I deactivate it, and I check manually later on
set.seed(123)
n_iterations <- 20
res <- bench::mark(check = FALSE,
"base::read.csv" = read.csv(temp_file_csv),
"arrow::read_csv_arrow" = arrow::read_csv_arrow(temp_file_csv),
"readr::read_csv" = readr::read_csv(
temp_file_csv,
show_col_types = FALSE,
progress = FALSE),
"data.table::fread" = data.table::fread(temp_file_csv),
"arrow::read_parquet" = arrow::read_parquet(temp_file_parquet),
iterations = n_iterations
)
Results
The winner is: data.table::fread!
data.table::fread and arrow::read_parquet
have comparable speed.
However, data.table consumes much more memory (5.87MB)
compared to arrow::read_parquet (1.08MB)
res %>%
select(expression, min, median, mem_alloc)
# A tibble: 5 x 4
expression min median mem_alloc
<bch:expr> <bch:tm> <bch:tm> <bch:byt>
1 base::read.csv 184ms 187.7ms 26.88MB
2 arrow::read_csv_arrow 30.4ms 32.2ms 5.36MB
3 readr::read_csv 139.6ms 143.1ms 1.45MB
4 data.table::fread 16.7ms 17.8ms 5.87MB
5 arrow::read_parquet 15.8ms 16.7ms 1.08MB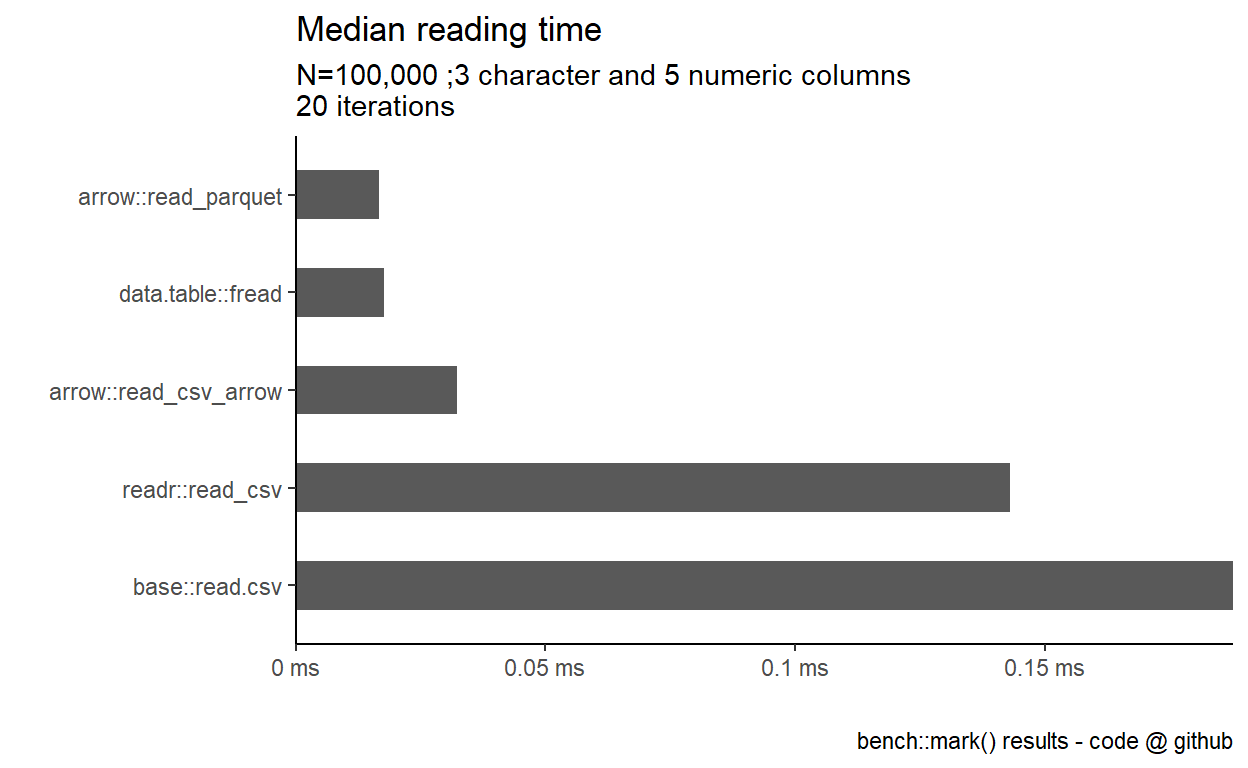
Size on disk
Significantly lower file size for the .parquet file
compared to the .csv.
.csv size: 4.86 MB.parquet size: 0.43 MBDoes the content matter?
A (rough) check whether the column type [characters or numbers] have any effect on the reading time.
I create 3 datasets; one with mostly numeric columns, one with mostly character columns and one with half numeric and half character columns
# borrowed from
# https://bench.r-lib.org/index.html#benchpress
create_numeric_df <- function(rows, cols) {
as.data.frame(setNames(
replicate(cols, runif(rows, 1, 1000), simplify = FALSE),
rep_len(c("x", letters), cols)))
}
create_character_df <- function(rows, cols) {
as.data.frame(setNames(
replicate(cols, sample(month.name, rows, replace = TRUE), simplify = FALSE),
rep_len(c("x", LETTERS), cols)))
}
n_cols <- 10; n_rows <- 100000
some_string <- c("a string here", "a string there")
# Mostly numeric. 9 numeric and 1 character
dta_numeric <- create_numeric_df(n_rows,n_cols)
dta_numeric$x = some_string
# Mostly character 9 character and 1 numeric
dta_character <- create_character_df(n_rows, n_cols)
dta_character$x <- runif(n_rows, 1, 1000)
# Mixed. 5 numeric - 5 character
dta_mixed <- bind_cols(
dta_numeric[letters[1:5]],
dta_character[LETTERS[1:5]]
)
types <- c("numeric", "character", "mixed") %>% set_names()
csv_files <- imap(types, ~ tempfile(., fileext = ".csv"))
parquet_files <- imap(types, ~tempfile(., fileext = ".parquet"))
# write the scv files
walk2(csv_files, names(csv_files), function(path, type) {
switch (type,
"numeric" = write.csv(dta_numeric, path, row.names = FALSE),
"character" = write.csv(dta_character, path, row.names = FALSE),
"mixed" = write.csv(dta_mixed, path, row.names = FALSE),
stop("No such type")
)
})
# write the .parquet files
walk2(parquet_files, names(parquet_files), function(path, type) {
switch (type,
"numeric" = arrow::write_parquet(dta_numeric, path),
"character" = arrow::write_parquet(dta_character, path),
"mixed" = arrow::write_parquet(dta_mixed, path),
stop("No such type")
)
})
Let’s run the comparison
I use the bench::press()
to run a grid of comparisons across the 3 datasets.
res_multi <- bench::press(
type = types,
{
set.seed(123)
n_iterations <- 20
res <- bench::mark(check = FALSE,
"base::read.csv" = read.csv(csv_files[[type]]),
"arrow::read_csv_arrow" = arrow::read_csv_arrow(csv_files[[type]]),
"readr::read_csv" = readr::read_csv(
csv_files[[type]],
show_col_= FALSE,
progress = FALSE),
"data.table::fread" = data.table::fread(csv_files[[type]]),
"arrow::read_parquet" = arrow::read_parquet(parquet_files[[type]]),
iterations = n_iterations
)
}
)
# don't forget to cleanup
walk(csv_files, unlink)
walk(parquet_files, unlink)
ggplot2::autoplot(res_multi, type = "violin")
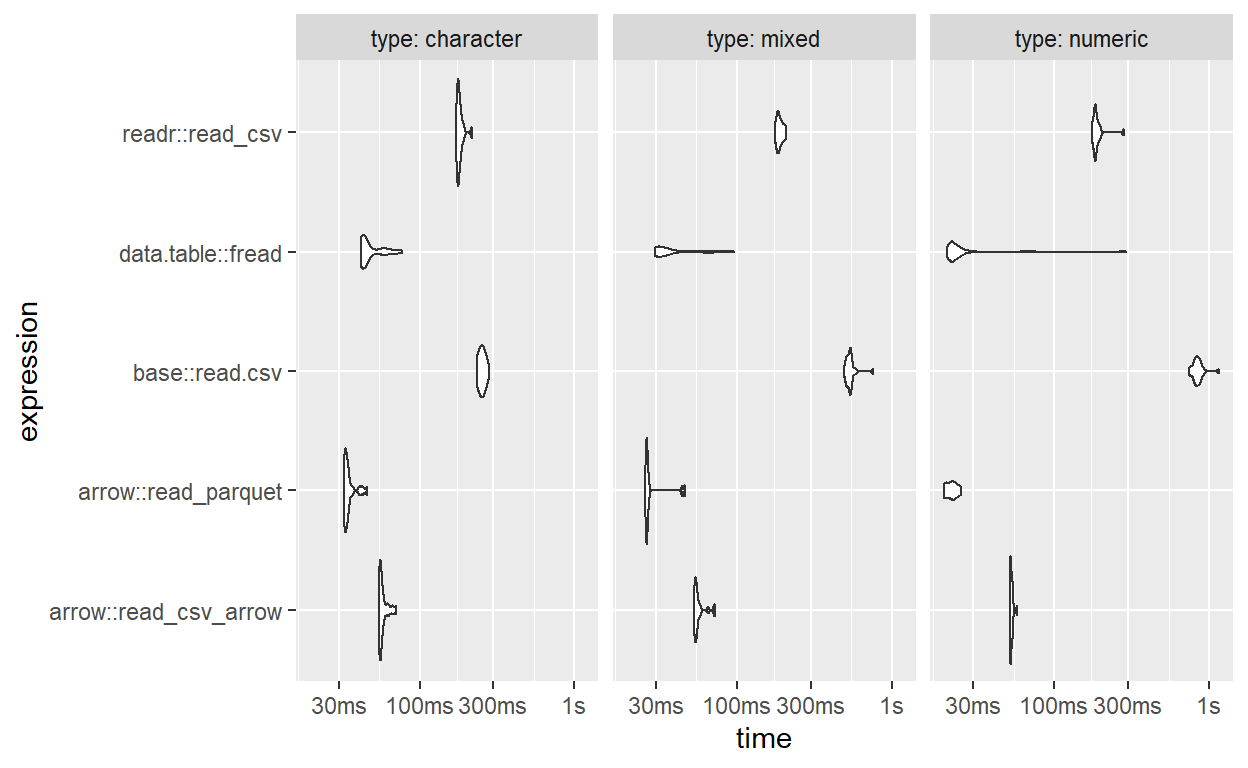
Median reading times
| Median reading times | |||
|---|---|---|---|
| 100,000 rows dataset | |||
| expression | Type of data | ||
| numeric | character | mixed | |
| arrow::read_parquet | 21.9ms | 34.0ms1 | 26.3ms1 |
| data.table::fread | 22.1ms2 | 43.5ms | 31.6ms |
| arrow::read_csv_arrow | 52.5ms | 56.1ms | 54.7ms |
| readr::read_csv | 184.4ms | 178.9ms | 186.5ms |
| base::read.csv | 767.4ms | 241.0ms | 505.6ms |
bench::mark() results - code @ github |
|||
|
1
The fastest in mostly character or mixed data
2
The fastest in mostly numeric data
|
|||
Same results, when reading the data files?
Let’s check that we get the same data back, when reading from disk
with all these readers. I did use the
bench::mark(check = TRUE) but it seems that when reading
with the readr::read_csv, I get the integer
columns flipper_length_mm, body_mass_g and
year back as double
readr::read_csv(temp_file_csv) %>% map_chr(typeof)
species island bill_length_mm
"character" "character" "double"
bill_depth_mm flipper_length_mm body_mass_g
"double" "double" "double"
sex year
"character" "double" whereas the other readers (e.g. data.table::fread),
return them as integer
data.table::fread(temp_file_csv)%>% map_chr(typeof)
species island bill_length_mm
"character" "character" "double"
bill_depth_mm flipper_length_mm body_mass_g
"double" "integer" "integer"
sex year
"character" "integer" When I explicitly asked the readr::read_csv to read them
as integer columns, then the only differences that remain
are in the class of the object returned.
csv_base = read.csv(temp_file_csv)
csv_arrow = arrow::read_csv_arrow(temp_file_csv)
csv_readr = readr::read_csv(temp_file_csv,
show_col_types = FALSE, progress = FALSE,
col_types = cols(
flipper_length_mm = col_integer(),
body_mass_g = col_integer(),
year = col_integer()
)
)
csv_data_table = data.table::fread(temp_file_csv)
parquet_arrow = arrow::read_parquet(temp_file_parquet)
readers <- c("csv_base","csv_readr", "csv_arrow", "csv_data_table", "parquet_arrow")
tbl_check <- expand.grid(
method1 = readers, method2 = readers,
stringsAsFactors = FALSE
) %>%
filter(method1 < method2) %>%
mutate(
comparison = map2(method1, method2, ~waldo::compare(
get(.x), get(.y),
x_arg = .x, y_arg = .y
)))
tbl_check$comparison
[[1]]
`class(csv_arrow)`: "tbl_df" "tbl" "data.frame"
`class(csv_base)`: "data.frame"
[[2]]
`class(csv_base)`: "data.frame"
`class(csv_readr)`: "spec_tbl_df" "tbl_df" "tbl" "data.frame"
[[3]]
`class(csv_arrow)`: "tbl_df" "tbl" "data.frame"
`class(csv_readr)`: "spec_tbl_df" "tbl_df" "tbl" "data.frame"
[[4]]
`class(csv_data_table)`: "data.table" "data.frame"
`class(csv_readr)`: "spec_tbl_df" "tbl_df" "tbl" "data.frame"
[[5]]
`class(csv_base)`: "data.frame"
`class(csv_data_table)`: "data.table" "data.frame"
[[6]]
`class(csv_arrow)`: "tbl_df" "tbl" "data.frame"
`class(csv_data_table)`: "data.table" "data.frame"
[[7]]
`class(csv_base)`: "data.frame"
`class(parquet_arrow)`: "tbl_df" "tbl" "data.frame"
[[8]]
`class(csv_readr)`: "spec_tbl_df" "tbl_df" "tbl" "data.frame"
`class(parquet_arrow)`: "tbl_df" "tbl" "data.frame"
[[9]]
v No differences
[[10]]
`class(csv_data_table)`: "data.table" "data.frame"
`class(parquet_arrow)`: "tbl_df" "tbl" "data.frame"Don’t forget to clean up people
That’s it! Hope you enjoyed it